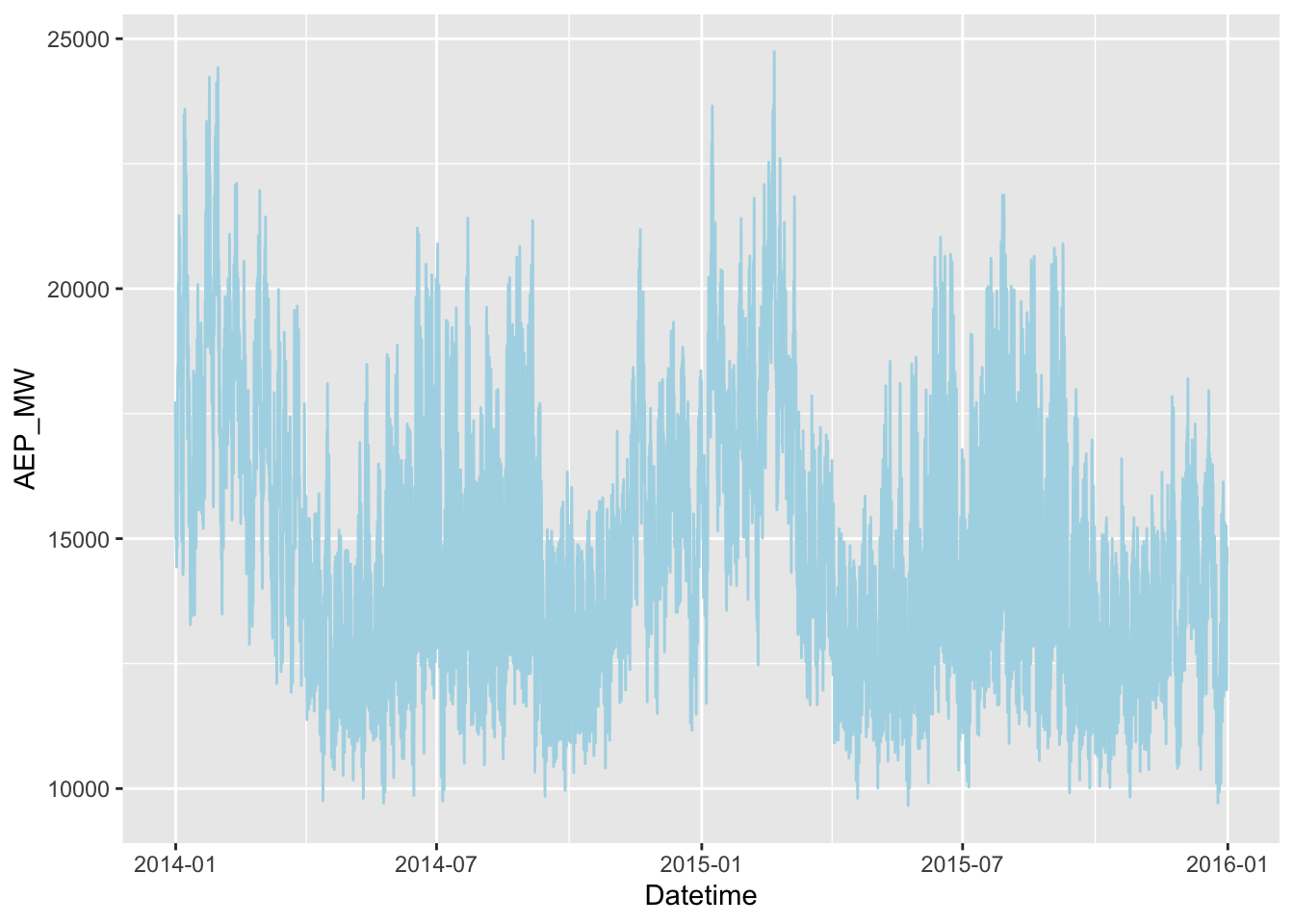
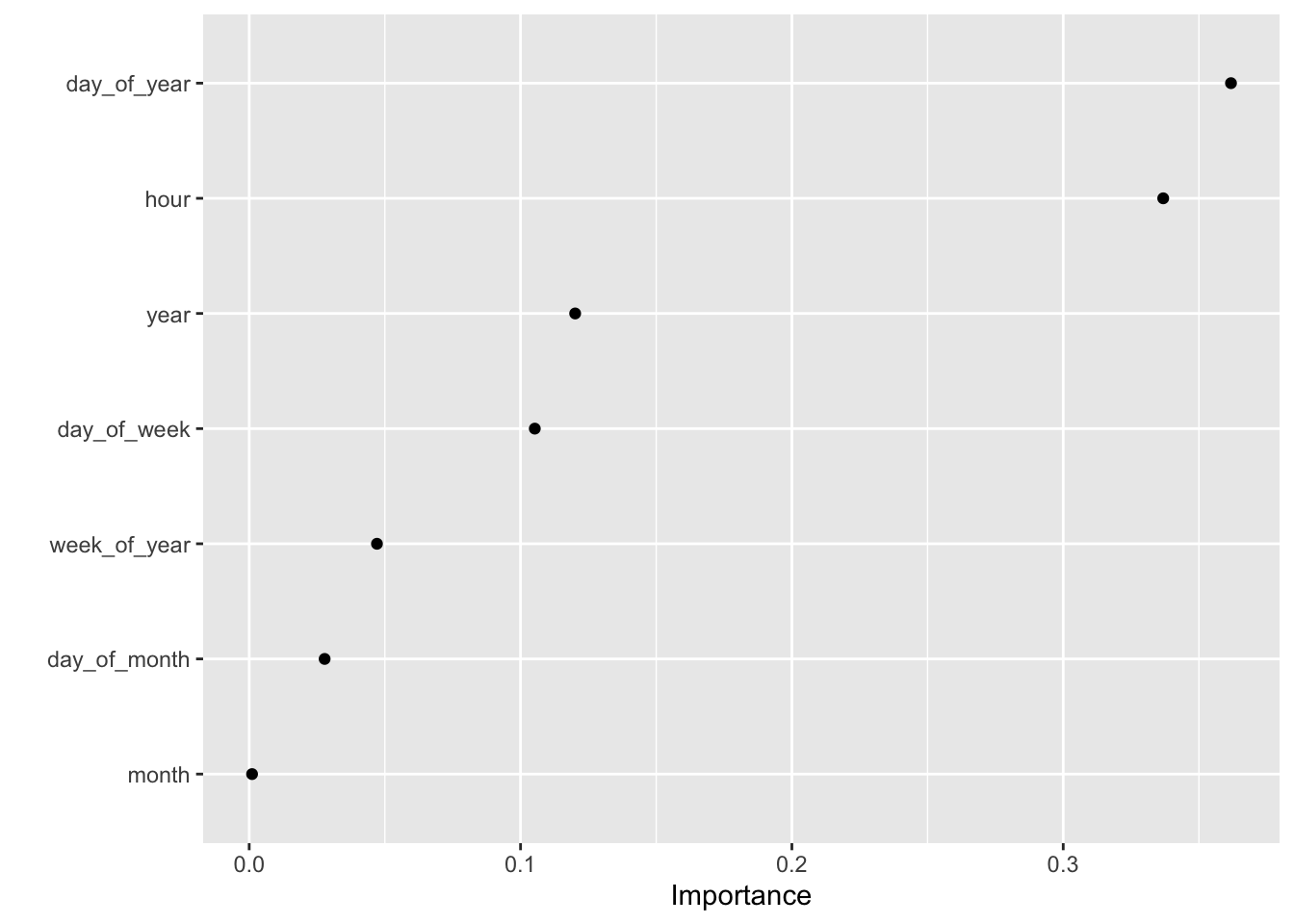
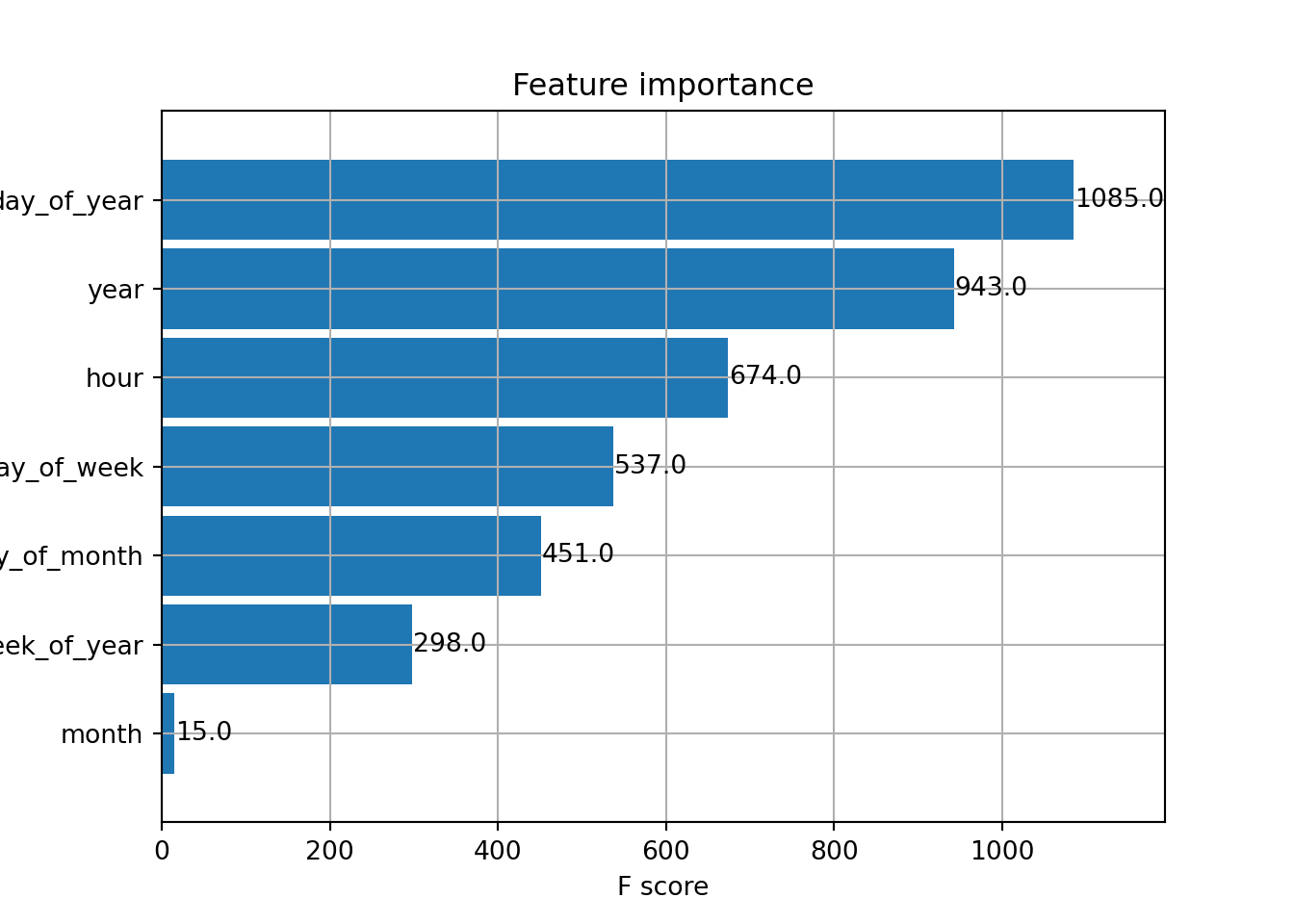
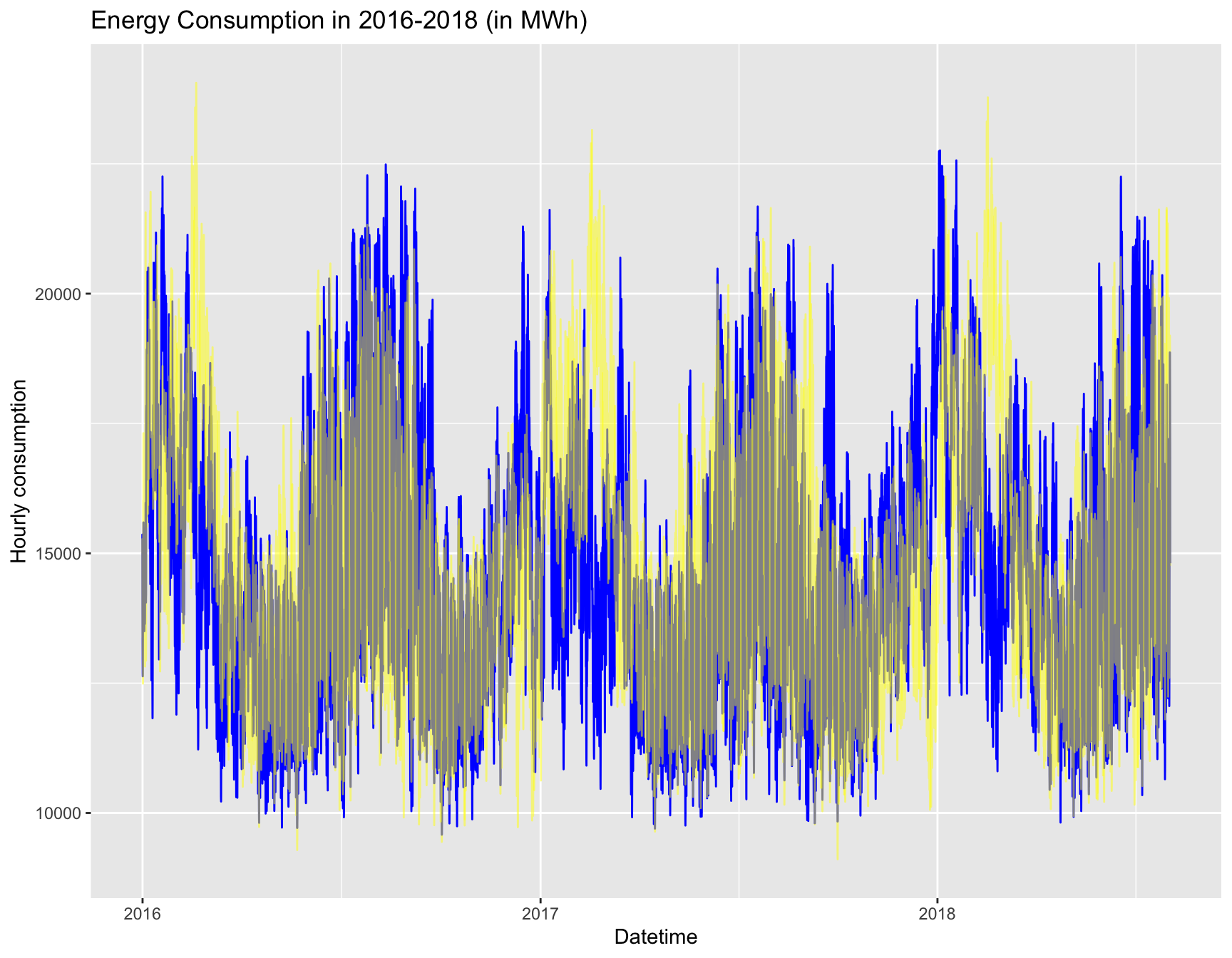
Rows: 121,273
Columns: 10
$ Datetime <dttm> 2004-12-31 01:00:00, 2004-12-31 02:00:00, 2004-12-31 03:…
$ AEP_MW <dbl> 13478, 12865, 12577, 12517, 12670, 13038, 13692, 14297, 1…
$ hour <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17…
$ day_of_week <dbl> 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, …
$ day_of_year <dbl> 366, 366, 366, 366, 366, 366, 366, 366, 366, 366, 366, 36…
$ day_of_month <int> 31, 31, 31, 31, 31, 31, 31, 31, 31, 31, 31, 31, 31, 31, 3…
$ week_of_year <dbl> 53, 53, 53, 53, 53, 53, 53, 53, 53, 53, 53, 53, 53, 53, 5…
$ month <dbl> 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 1…
$ quarter <int> 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, …
$ year <dbl> 2004, 2004, 2004, 2004, 2004, 2004, 2004, 2004, 2004, 200…